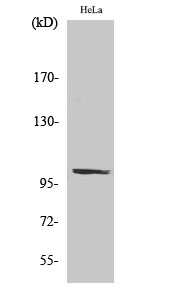
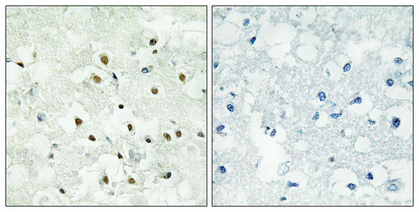
PMS2 Polyclonal Antibody
- 产品详情
- 实验流程
- 背景知识
Application
| WB, IHC-P, IF, ICC, E |
|---|---|
| Primary Accession | P54278 |
| Reactivity | Human, Rat, Mouse |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 95797 Da |
| Gene ID | 5395 |
|---|---|
| Other Names | PMS2; PMSL2; Mismatch repair endonuclease PMS2; DNA mismatch repair protein PMS2; PMS1 protein homolog 2 |
| Dilution | WB~~Western Blot: 1/500 - 1/2000. Immunohistochemistry: 1/100 - 1/300. Immunofluorescence: 1/200 - 1/1000. ELISA: 1/10000. Not yet tested in other applications. IHC-P~~Western Blot: 1/500 - 1/2000. Immunohistochemistry: 1/100 - 1/300. Immunofluorescence: 1/200 - 1/1000. ELISA: 1/10000. Not yet tested in other applications. IF~~1:50~200 ICC~~N/A E~~N/A |
| Format | Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.09% (W/V) sodium azide. |
| Storage Conditions | -20℃ |
| Name | PMS2 (HGNC:9122) |
|---|---|
| Function | Component of the post-replicative DNA mismatch repair system (MMR) (PubMed:30653781, PubMed:35189042). Heterodimerizes with MLH1 to form MutL alpha. DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages. Possesses an ATPase activity, but in the absence of gross structural changes, ATP hydrolysis may not be necessary for proficient mismatch repair (PubMed:35189042). |
| Cellular Location | Nucleus |
For Research Use Only. Not For Use In Diagnostic Procedures.
Provided below are standard protocols that you may find useful for product applications.
BACKGROUND
Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MLH1 to form MutL alpha. DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2- MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages.
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.























 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。