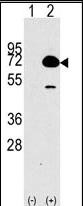
HDAC10 Antibody (N-term)
Purified Rabbit Polyclonal Antibody (Pab)
- 产品详情
- 文献引用 : 1
- 实验流程
- 背景知识
Application
| WB, E |
|---|---|
| Primary Accession | Q969S8 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 71445 Da |
| Antigen Region | 16-46 aa |
| Gene ID | 83933 |
|---|---|
| Other Names | Histone deacetylase 10, HD10, HDAC10 |
| Target/Specificity | This HDAC10 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 16-46 amino acids from the N-terminal region of human HDAC10. |
| Dilution | WB~~1:1000 E~~Use at an assay dependent concentration. |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | HDAC10 Antibody (N-term) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | HDAC10 |
|---|---|
| Function | Polyamine deacetylase (PDAC), which acts preferentially on N(8)-acetylspermidine, and also on acetylcadaverine and acetylputrescine (PubMed:28516954). Exhibits attenuated catalytic activity toward N(1),N(8)-diacetylspermidine and very low activity, if any, toward N(1)-acetylspermidine (PubMed:28516954). Histone deacetylase activity has been observed in vitro (PubMed:11677242, PubMed:11726666, PubMed:11739383, PubMed:11861901). Has also been shown to be involved in MSH2 deacetylation (PubMed:26221039). The physiological relevance of protein/histone deacetylase activity is unclear and could be very weak (PubMed:28516954). May play a role in the promotion of late stages of autophagy, possibly autophagosome- lysosome fusion and/or lysosomal exocytosis in neuroblastoma cells (PubMed:23801752, PubMed:29968769). May play a role in homologous recombination (PubMed:21247901). May promote DNA mismatch repair (PubMed:26221039). |
| Cellular Location | Cytoplasm. Nucleus Note=Excluded from nucleoli. |
| Tissue Location | Widely expressed with high levels in liver and kidney. |
For Research Use Only. Not For Use In Diagnostic Procedures.

Provided below are standard protocols that you may find useful for product applications.
BACKGROUND
Histone deacetylase (HDAC) and histone acetyltransferase (HAT) are enzymes that regulate transcription by selectively deacetylating or acetylating the eta-amino groups of lysines located near the amino termini of core histone proteins (1). Eight members of HDAC family have been identified in the past several years (2,3). These HDAC family members are divided into two classes, I and II. Class I of the HDAC family comprises four members, HDAC-1, 2, 3, and 8, each of which contains a deacetylase domain exhibiting from 45 to 93% identity in amino acid sequence. Class II of the HDAC family comprises HDAC-4, 5, 6, and 7, the molecular weights of which are all about two-fold larger than those of the class I members, and the deacetylase domains are present within the C-terminal regions, except that HDAC-6 contains two copies of the domain, one within each of the N-terminal and C-terminal regions. Human HDAC-1, 2 and 3 were expressed in various tissues, but the others (HDAC-4, 5, 6, and 7) showed tissue-specific expression patterns (3). These results suggested that each member of the HDAC family exhibits a different, individual substrate specificity and function in vivo. HDAC8 interacts with PEPB2-MYH11, a fusion protein consisting of the 165 N-terminal residues of CBF-beta (PEPB2) with the tail region of MYH11 produced by the inversion Inv(16)(p13q22), a translocation associated with acute myeloid leukemia of M4EO subtype. The PEPB2-MYH1 fusion protein also interacts with RUNX1, a well known transcriptional regulator, suggesting that the interaction with HDAC8 may participate to convert RUNX1 into a constitutive transcriptional repressor.
REFERENCES
Keedy, K.S. et al. J Virol. May; 83(10): 4749?756(2009).
Tong, J.J., et al., Nucleic Acids Res. 30(5):1114-1123 (2002).
Fischer, D.D., et al., J. Biol. Chem. 277(8):6656-6666 (2002).
Guardiola, A.R., et al., J. Biol. Chem. 277(5):3350-3356 (2002).
Kao, H.Y., et al., J. Biol. Chem. 277(1):187-193 (2002).
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.






















 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。